A Comprehensive Benchmark on 2D/3D Classification and Segmentation
- Impressive Transferability: DINOv3, trained solely on natural images, delivers outstanding performance in medical vision tasks like CT and X-ray analysis, often surpassing specialized models without any domain-specific tweaks.
- Key Limitations Exposed: The model struggles in highly specialized areas such as Whole-Slide Pathological Images (WSI), Electron Microscopy (EM), and Positron Emission Tomography (PET), highlighting gaps in handling extreme domain shifts.
- Scaling Laws Challenged: Unlike in natural image tasks, larger model sizes and higher resolutions don’t consistently boost performance in medical imaging, revealing task-dependent behaviors and opening doors for future innovations like multiview 3D reconstruction.
In the ever-evolving world of computer vision, large-scale foundation models have transformed how machines “see” the world, drawing inspiration from the success of Large Language Models (LLMs) that learn vast knowledge from unannotated data. These vision models, pre-trained on billions of diverse natural images, promise to generalize across domains—but what happens when we apply them to the intricate, high-stakes field of medical imaging? This is the core question driving a groundbreaking benchmark of DINOv3, the latest in a series of self-supervised vision transformers (ViTs) that have pushed the boundaries of visual representation learning. Developed as part of the DINO lineage, DINOv3 scales up to an astonishing 7 billion parameters trained on 1.7 billion images, excelling in dense prediction tasks like segmentation and classification. Yet, its true test lies in whether this powerhouse, honed on everyday photos, can tackle the unique challenges of medical visuals without any fine-tuning or medical-specific pre-training.
Medical imaging isn’t just about pretty pictures—it’s a lifeline for diagnosing diseases, from subtle anomalies in X-rays to complex 3D structures in CT scans. The field grapples with a dizzying array of modalities: 2D grayscale images like chest X-rays, multi-channel RGB histopathology slides, and volumetric 3D scans from MRI or CT machines. Add to that long-tailed distributions of rare conditions, prohibitive data collection costs, and strict privacy regulations, and you have a domain crying out for robust, off-the-shelf tools. Traditional approaches often rely on domain-specific models like BiomedCLIP, which trains on web-crawled medical images with text supervision, or CT-Net, tailored for 3D tasks. But what if a generalist like DINOv3 could step in? This benchmark puts it to the test across a wide spectrum of tasks, including 2D and 3D classification and segmentation, systematically varying model sizes and input resolutions to probe its scalability.
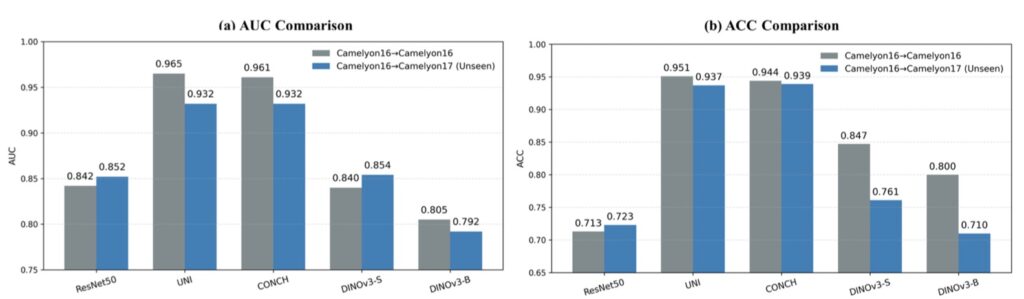
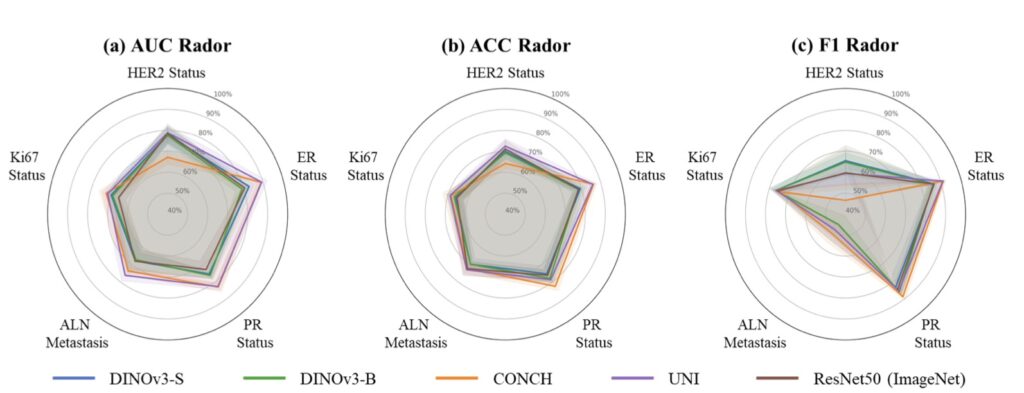
The results are nothing short of eye-opening. DINOv3 shines brightly in areas with visual similarities to natural images, such as CT and X-ray analysis, where it establishes a formidable new baseline. Remarkably, it outperforms medical-specific heavyweights like BiomedCLIP and CT-Net on several benchmarks, all while being trained exclusively on non-medical data. This suggests that the rich, emergent representations from self-supervised learning (SSL) on massive natural image datasets can transfer powerfully to medical tasks, providing a robust prior for spotting patterns like tumors or fractures. Recent studies have hinted at this potential, showing promising results with DINOv3 on niche medical applications, but this comprehensive evaluation clarifies its broader impact, proving it’s not just a fluke but a scalable strength.
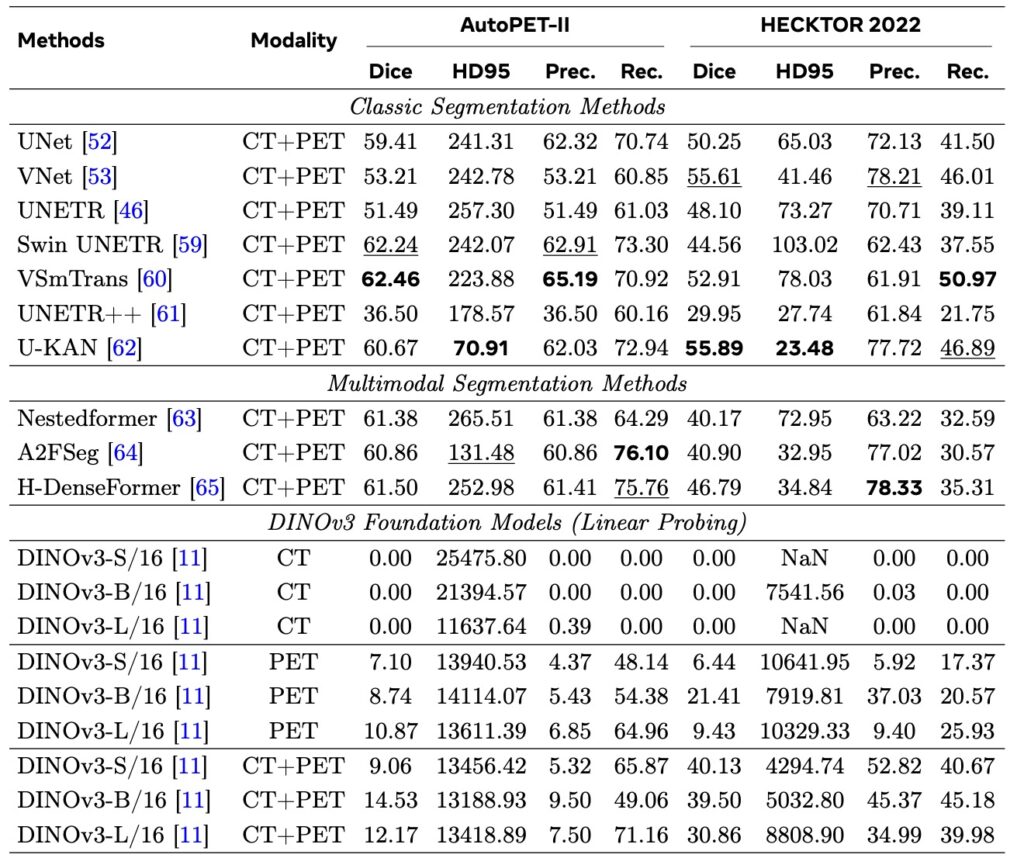
However, no model is perfect, and DINOv3’s limitations paint a nuanced picture of its domain transfer. Performance takes a nosedive in ultra-specialized scenarios requiring deep domain knowledge, such as Whole-Slide Pathological Images (WSIs) with their gigapixel resolutions, Electron Microscopy (EM) capturing cellular intricacies, and Positron Emission Tomography (PET) dealing with functional imaging. Here, the gap between natural image training and these extreme distributions becomes glaring, leading to degraded features and suboptimal outcomes. It’s a reminder that while DINOv3’s SSL roots—building on predecessors like DINOv1 and DINOv2—excel in generalization, they can’t fully bridge every chasm without some adaptation. This echoes broader debates in vision research, where scaling laws (the idea that bigger models and more data yield predictable gains) have been assumed but not always proven, especially beyond narrow tasks.
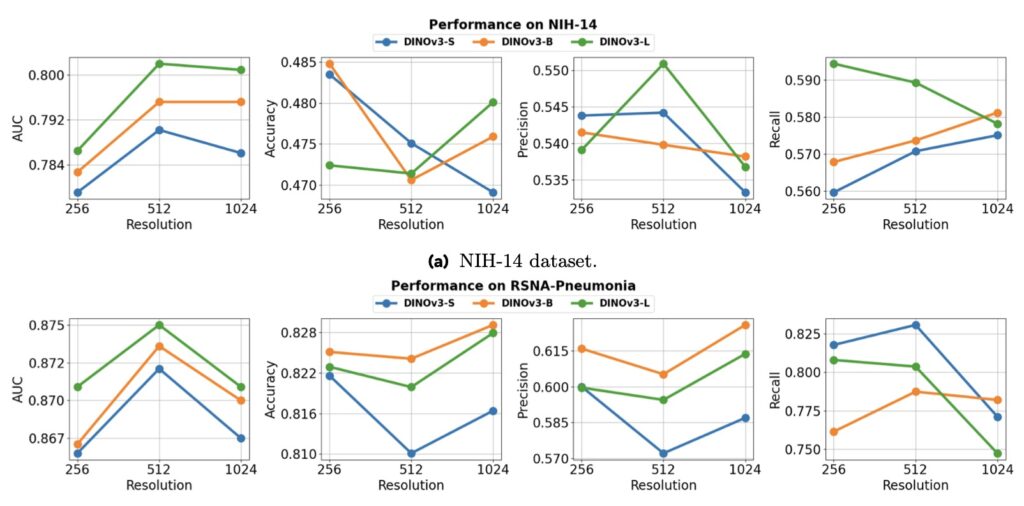
Speaking of scaling, this benchmark dives deep into how DINOv3 behaves when you crank up the dials. In the natural image realm, the paradigm of large-scale modeling has led to impressive scaling laws, as seen in LLMs and earlier vision works. DINOv3 itself demonstrates this on general tasks, but in medicine? Not so straightforward. Our analysis reveals that performance doesn’t reliably climb with larger models or finer feature resolutions; instead, it shows diverse, task-dependent behaviors. For instance, a bigger model might boost segmentation accuracy in 3D CT volumes but falter in 2D histopathology, defying the “bigger is always better” mantra. This challenges assumptions from prior research, which often focused on limited evaluations, and underscores the medical domain’s heterogeneity as a tough proving ground for visual pre-training.
This report positions DINOv3 as more than just a tool—it’s a catalyst for the future of medical vision. By serving as a strong, unified encoder without needing costly medical data curation, it paves the way for innovations like enforcing multiview consistency in 3D reconstruction, potentially revolutionizing how we rebuild anatomical structures from scans. While efforts like BiomedCLIP address data scarcity through clever web-scraping and supervision, DINOv3 proves that natural image foundations can be a surprisingly effective starting point. For researchers and practitioners, this opens exciting directions: fine-tuning with minimal medical data, hybrid models blending generalist and specialist strengths, or even scaling up medical-specific datasets to match natural image volumes. As computer vision continues to borrow from the LLM playbook, DINOv3’s benchmark reminds us that the path to a true “medical vision foundation” is promising but paved with domain-specific hurdles. Will it set the new standard? The evidence says it’s close, but the journey is just beginning.
